Project Overview
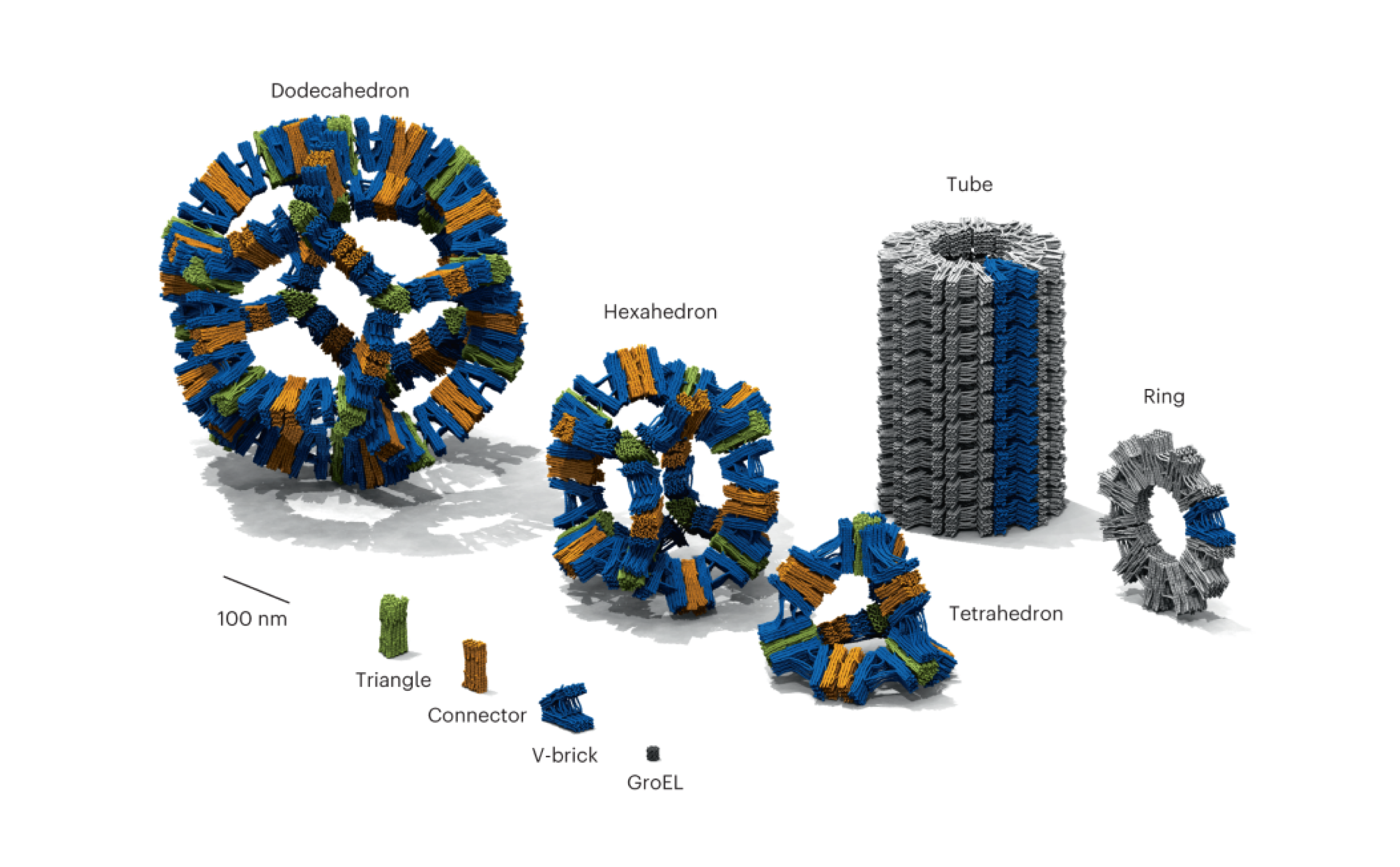
DeepSNUPI represents a breakthrough in computational biology, utilizing graph neural networks to predict DNA origami shapes with unprecedented accuracy. This innovative approach enables rational design of complex nanostructures, opening new possibilities for programmable molecular systems.
Goals
- Accurate Shape Prediction: Develop machine learning models that can reliably predict the 3D structure of DNA origami designs
- Rational Design Tools: Create computational frameworks for designing DNA origami with desired properties
- Scalable Methodology: Build approaches that can handle increasingly complex nanostructure designs
- Experimental Validation: Bridge computational predictions with experimental verification
Key Achievements
- Published in Nature Materials (2024) - one of the most prestigious journals in materials science
- Developed novel graph neural network architecture specifically for DNA origami structures
- Achieved state-of-the-art prediction accuracy for complex 3D nanostructures
- Created open-source tools for the DNA nanotechnology community
Team Members
Principal Investigator
- Prof. Do-Nyun Kim - Seoul National University
Collaborators
- Dr. Chien Truong-Quoc - Co-Lead, Algorithm Development
- Dr. Jae Young Lee - Co-Lead, Algorithm Development
- Dr. Kyung Soo Kim - Experimental validation
Students
- Graduate students and undergraduate researchers contributing to ongoing developments
Publications
- Truong-Quoc, C., Lee, J.Y., Kim, K.S., Kim, D.N. “Prediction of DNA origami shape using graph neural network.” Nature Materials (2024). DOI: 10.1038/s41563-024-01846-8
Future Directions
- Extension to more complex DNA origami architectures
- Integration with experimental design workflows
- Development of real-time prediction tools
- Applications in drug delivery and biosensing systems